 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Gene information Gene information |
|||
| Locus | FGSG_01182 | Length | 277 aa |
| Gene Name | GzCCAAT002 | # Motifs | 1 |
| Family-1 | Heteromeric CCAAT factors | Deletion | Y |
| Family-2 | Parent strain | GZ3639 | |
| Family-3 | Reference | ||
 Primers Primers |
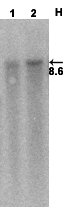
S o u t h e r n b l o t |
 |
|
| name | sequence | ||
| 5F | GGGAGGCCTTTCACCACGAC | ||
| 5R | gcacaggtacacttgtttagagCTTTTGCGCGAGCTGGATTTAC | ||
| 3F | ccttcaatatcatcttctgtcgAGGAAGGAAAGGAAGACAGAGACGAG | ||
| 3R | TCACCAACACAGTCGAACATCAGG | ||
| 5N | GAGCATGCGTACCCCAGTATCCAT | ||
| 3N | GGTGGTGATACGAAATGACGGAATG | ||
| with 5F | CGGGGATGGTCTCAGCACTTATG | ||
 Phenotypes Phenotypes |
Wild type | Mutant | |
| Mycelia growth | PDA | 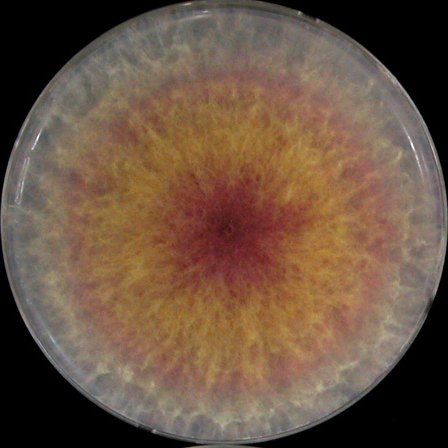 |
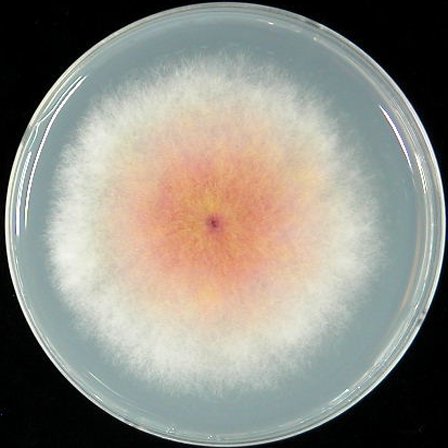 |
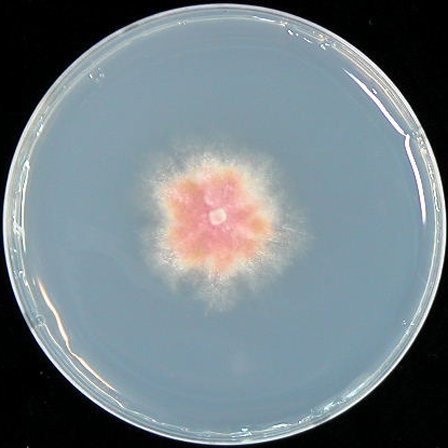
| MM | 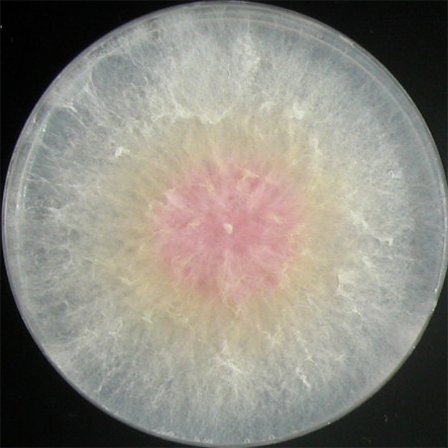 |
 |
|
| Sexual development | Number of perithecia (NP) | 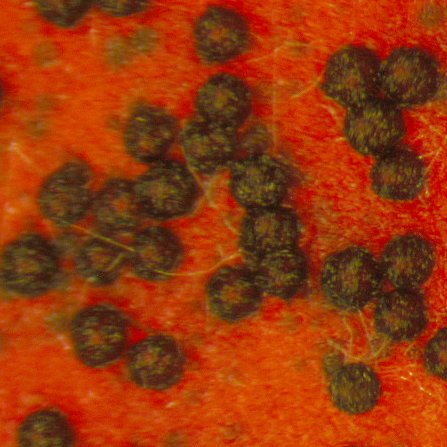 |
 |
| Perithecia maturation (PM) | |||
| Ascospore formation (AF) | 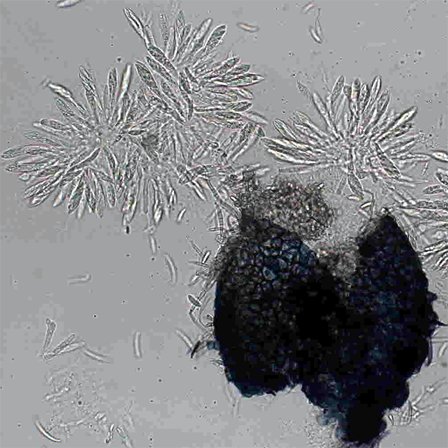 |
 |
|
| Ascospore discharge (AD) | |||
| Conidiation (106) | 0.24 | 0.06 | |
| Toxin Production | No image | No image | |
| Virulence |  |
 |
|
| Stress responses | |||
 |
|||